Introduction
Antibiotics have been used as effective drugs to treat infectious diseases in humans and animals. However, after the introduction of antibiotics, appearance of multidrug-resistant bacteria, which showed resistance to various antibiotics, increased. This is emerging as a social problem because of the economic loss and deaths caused by the difficulty in treatment of diseases of humans and animals. In the livestock sector, the use of antibiotics was restricted to establish countermeasures of antibiotic resistant bacteria, and in particular, was prohibited in formula feeds. Recently, interest has been increasing for the development of therapeutic agents to treat various diseases using natural resources (Son et al., 2018). In addition, efforts to prevent disease and maintain optimal health using natural supplements via preventative medicine are increasing. Studies assessing the anticancer effects of targeted natural substances have been performed, resulting in the identification of substances that efficiently promote anticancer effects and boost the immune system; additional investigations extracting natural products extraction are actively being conducted. Lipopolysaccharide (LPS), a common component of the cell walls of Gram-negative bacteria, is an endotoxin that causes endotoxin shock, tissue injury, and lethality (Walker, 1992). The immunological response to Gram-negative bacteria mainly involves leukocytes and the production of cytokines such as tumor necrosis factor alpha (TNF-α) and interleukin-1 (IL-1) (Van Amersfoort et al., 2003). The inflammatory cascade triggered by these cytokines eventually causes toxic shock and death (Luheshi and Rothwell, 1996; Gunther et al., 2011). Concanavalin-A (Con-A) is both a plant and lymphocyte mitogen. It stimulates T-cell subsets to give rise to four functionally distinct T-cell populations (Gantner et al., 1995).
Osthole [7-methoxy-8-(3-methyl-but-2-enyl)-chromen-2-one] is an O-methylated natural coumarin and a fragrant organic chemical compound. It is a calcium channel blocker that is found in various plants including Cnidium monnieri, Angelica archangelica, and Angelica pubescens. It is used as a pharmacological, biological, and therapeutic agent throughout China (Teng et al., 1994; Hoult and Paya, 1996); it inhibits the release of inflammatory mediators (Nakamura et al., 2009) and displays many other pharmacological and biological effects (Okamoto et al., 2007; Tang et al., 2008).
Regulating pro-inflammatory cytokines is of great medical interest for the treatment of several inflammatory diseases, including asthma, autoimmune diseases, pelvic inflammatory disease, and sepsis. Sepsis and septic shock are serious, life-threatening disorders that are triggered by systemic infection and are characterized by the hypoperfusion of major organs, which leads to lethal multiple organ failure (MOF), shock, and death (Jung et al., 2008). The peripheral blood mononuclear cells (PBMCs), which are the major components of the immune system, can lead to protect the individual by performing an immune response from various infection. However, whether osthole exerts effects on PBMCs in Hanwoo cattle has remained unknown. In this study, the anti-inflammatory effects of osthole were investigated in PBMCs isolated from Hanwoo cattle.
Material and methods
Animals and materials
Blood samples used to isolate PBMCs were obtained from bulls at the National Institute of Animal Science (Suwon, Korea). Animals received care in accordance with the standard guideline for the Care and Use of Laboratory Animals provided by the National Institute of Animal Science Animal Care Committee, and the experiment was conducted with approval from the animal ethics committee and Operation rule of animal experiment ethics in the National Institute of Animal Science (approval number: 2014-083). Osthole, LPS, and Con-A were purchased from Sigma-Aldrich (St. Louis, USA). Cell culture media (RPMI-1640), fetal bovine serum (FBS), and penicillin/streptomycin were obtained from Gibco/BRL Life Technologies Inc. (Grand Island, NY, USA). All other materials were purchased from Sigma-Aldrich.
Cell isolation, culture and treatment
Blood sampling was performed in 18-month-old animals. Samples were collected into dipotassium–EDTA tubes, and PBMCs were then isolated by Ficoll-Paque density gradient centrifugation for 30 min at 1,200 × g at 25℃. The cells were washed three times with phosphate-buffered saline (PBS) and then incubated in RPMI-1640 medium supplemented with 10% heat-inactivated FBS, 1% glutamax-1, 100 IU/mL penicillin, and 100 μg/mL streptomycin at 37℃ in a 5% CO2 humidified air environment. For experiments, 1 × 106 cells were incubated for 6 h and 72 h in culture medium supplemented with 15 μg/mL LPS from Escherichia coli O111:B4 or 5 μg/mL of Con-A. The LPS and Con-A treatment refer to concentration that causes the highest stimulation of PBMCs (Adams and Czuprynski, 1990; Panadero et al., 2009). And optimal concentrations were determined in preliminary assays. PBMCs were then centrifuged for 10 min at 500 × g and harvested for RNA extraction. Supernatants were frozen at - 20℃ for cytokine quantification using an enzyme-linked immunosorbent assay (ELISA).
Cell viability assays
Cell viability was evaluated using the EZ-Cytox cell viability assay kit (Daeil Lab, Seoul, Korea) following the manufacturer’s instructions. Briefly, 10 μL of EZ-Cytox reagent was added to each well of 96-well plate and incubated for 2 h at 37℃. Plates were then agitated thoroughly for 1 min on an orbital shaker, and the optical density at 450 nm was measured using an ELISA plate reader (Versa max Microplate Reader; Molecular Devices, Sunnyvale, USA). Wells lacking cells but containing complete medium and EZ-Cytox reagent were used as blanks. All assays were performed in triplicate and expressed as the percentage of cell viability relative to untreated control cells.
RNA isolation and cDNA synthesis
Total RNA was extracted from cells using 1 mL of TRIzol® reagent (Invitrogen, Carlsbad, USA) according to the manufacturer’s protocol, as described previously (Nagai et al., 2005). RNA quality was confirmed by visualizing the 28 S and 18 S rRNA bands on 1.5% agarose gels stained with ethidium bromide. Total RNA was purified from all samples using an RNeasy MinElute cleanup kit (Qiagen, Valencia, USA) and quantified using a Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, USA).
Complementary DNA (cDNA) synthesis was performed by reverse transcription using SuperScript™ II reverse transcriptase (Invitrogen). Briefly, 4-μL aliquots of total RNA were preincubated with 50 ng (1 μL) of Oligo(dT) (Promega, Madison, USA) and 2.5 mM (1 μL) dNTP mix at 65℃ for 5 min. The tubes were placed on ice, and 4 μL of 5X first-stand buffer [250 mM Tris–HCl (pH 8.3), 375 mM KCl, 15 mM MgCl2], 2 μL of 0.1 M dithiothreitol (DTT), and 40 U (0.5 μL) of RNase inhibitor (Promega) were added, followed by incubation at 42℃ for 2 min. Then, 200 units (1 μL) of SuperScript™ II reverse transcriptase (Invitrogen) was added, and tubes were incubated at 42℃ for 50 min. The reverse transcriptase activity was terminated by incubation at 70℃ for 15 min. The resulting cDNA was stored at - 20℃ until use for quantitative real-time-polymerase chain reaction (qPCR) experiments.
Quantitative real-time polymerase chain reaction (qPCR)
qPCR was performed using the Power SYBR green PCR master mix and an ABI 7500 real-time PCR system (both from Applied Biosystems, Foster City, USA). All primer sets were designed using the Primer3 program (http://bioinfo.ut.ee/primer3-0.4.0) to amplify products ranging from 100 to 200 base pairs (Table 1). The glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene (GenBank Accession Number NM_001034034.2) was used as the internal control. The qPCR reactions were performed in a volume of 20 μL, including 2 μL of cDNA (0.1 μg/μL), 10 μL of 2X SYBR® Green PCR Master Mix (Applied Biosystems), and 1 μL of 10 pM forward and reverse primers. The amplification reaction was started by incubation for 2 min at 50℃, followed by 40 cycles at 95℃ for 10 min, 95℃ for 10 s, and 60℃ for 1 min, with a final extension step at 72℃ for 1 min. The qPCR reaction for each gene was repeated three times. Following amplification, melting curve analyses were performed to verify the specificity of the reactions. The endpoint used for real-time PCR quantification (Ct) was defined as the PCR threshold cycle number. The ΔCt was determined by subtracting the GAPDH Ct value for each sample from the target Ct value. Finally, the expression level was transformed to a 2–ΔCt value for further analysis. GAPDH was selected as the internal reference gene, and the 2–∆∆Ct method was used to calculate the fold changes in gene expression (Livak and Schmittgen, 2001).
|
Table 1. Primer list for quantitative Real Time-PCR. 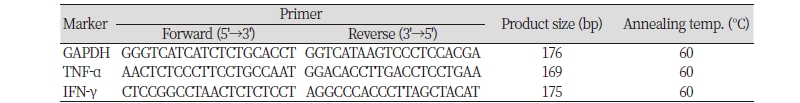
|
|
|
PCR, polymerase chain reaction; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; TNF-α, tumor necrosis factor-α; IFN-γ, interferon-γ. |
|
Enzyme-linked immunosorbent assay (ELISA)
The concentrations of TNF-α and IFN-γ were determined using commercially available ELISA kits (Bethyl Laboratories, Montgomery, USA) according to the manufacturer’s instructions. ELISA results were quantified using an ELISA plate reader at a wavelength of 450 nm. Results were reported as the mean values of duplicate ELISA wells.
Statistical analysis
GraphPad Prism software (GraphPad Software, San Diego, USA) was used for all statistical analyses and graphical presentation. All experiments were repeated at least three times. Data were expressed as means± SEM. Groups were compared using two-tailed parametric one-way ANOVA, and p values were adjusted for repeated testing by Bonferroni’s multiple comparison test. A statistic difference was accepted as significant when p < 0.05.
Results
Effects of osthole on PBMC viability
To assess the possibility that inhibited pro-inflammatory cytokine production was due to the cytotoxic effects of osthole, we performed EZ-Cytox assays in osthole-treated PBMCs. Doses of osthole ranging from 0.01 to 100 μM did not affect cell viability (Fig. 1), even at the highest concentration. Therefore, different dosages of osthole (0.1, 1, 10, and 100 μM) were used to evaluate the inhibitory effects of osthole on the LPS- or Con-A-induced pro-inflammatory cytokine (TNF-α and IFN-γ) production in PBMCs isolated from Hanwoo cattle.
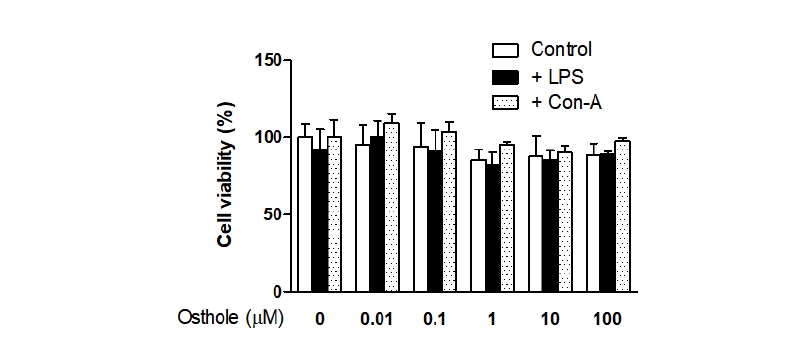
Fig. 1. Effects of osthole on peripheral blood mononuclear cells (PBMC) viability. Cells were treated with the indicated concentrations of osthole (0, 0.01, 0.1, 1, 10, and 100 μM) with lipopolysaccharide (LPS) (15 μ g/mL) or concanavalin-A (Con-A) (5 μg/mL) for 72 hours and cell viability was determined. Cell viability was assessed by EZ-Cytox assays, and the results were expressed as the percentage of surviving cells over control cells, respectively. Results indicated the mean ± SEM of three independent experiments.
Effects of osthole on LPS- or Con-A-induced TNF-α and IFN-γ production
Because TNF-α and IFN-γ are important cytokines during inflammatory reactions, we evaluated the effects of osthole on LPS- or Con-A-induced TNF-α and IFN-γ production by ELISA. Treatment with LPS increased the production of TNF-α compared to the control (Fig. 2A). Conversely, treatment with osthole significantly inhibited LPS-induced TNF-α in a dose-dependent manner compared to LPS-treated cells. Osthole showed significant inhibitory effects at a dose of 0.1 μM (p < 0.05). Next, we measured LPS-induced IFN-γ production to confirm the inhibitory effects of osthole (Fig. 2B). However, treatment with LPS did not cause a significant difference in IFN-γ production compared to the control or osthole-treated groups.
Treatment with Con-A increased the production of TNF-α compared to the control (Fig. 3A). In contrast, treatment with osthole significantly inhibited Con-A-induced TNF-α production in a dose-dependent manner compared to Con-A-treated cells. Osthole inhibited Con-A-induced TNF-α at a dose of 10 μM (p < 0.05). We next measured Con-A-induced IFN-γ production to confirm the inhibitory effects of osthole (Fig. 3B). Treatment with Con-A induced IFN-γ production significantly compared to the osthole-treated group; osthole inhibited Con-A-induced IFN-γ at a dose of 10 μM (p < 0.01). Importantly, the highest dose of osthole inhibited the effects of LPS or Con-A entirely. These results confirm the inhibitory effects of osthole on TNF-α and IFN-γ activity in PBMCs.
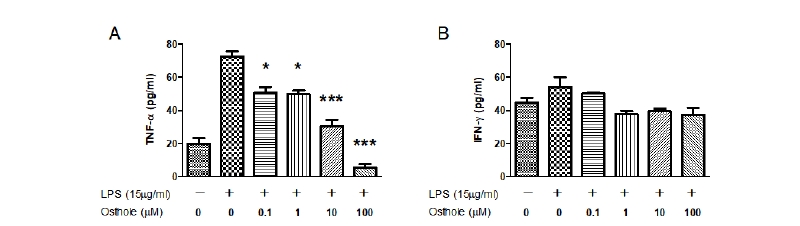
Fig. 2. Effects of osthole on lipopolysaccharide (LPS)-induced tumor necrosis factor-α (TNF-α) and interferon-γ (IFN-γ) production in peripheral blood mononuclear cells (PBMC). Cells were treated with the indicated concentrations of osthole (0, 0.1, 1, 10, and 100 μM) with LPS (15 μg/mL) for 72 hours, respectively. Treatment with osthole significantly inhibited LPS-induced TNF-α in a dose-dependent manner compared to LPStreated cells (A). Treatment with LPS did not cause a significant difference in IFN-γ production compared to the control or osthole-treated groups (B). The extracellular levels of TNF-α and IFN-γ were measured in culture media using commercial ELISA kits. Results indicated the mean ± SEM of three independent experiments. The asterisks show statistically significant values (*p < 0.05 and ***p < 0.001) and compared with only LPS treated group
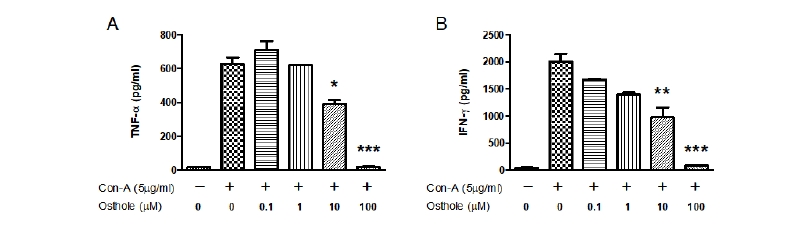
Fig. 3. Effects of osthole on concanavalin-A (Con-A)-induced tumor necrosis factor-α (TNF-α) and interferon-γ (IFN-γ) production in peripheral blood mononuclear cells (PBMC). Cells were treated with the indicated concentrations of osthole (0, 0.1, 1, 10, and 100 μM) with Con-A (5 μg/mL) for 72 hours, respectively. Treatment with osthole significantly inhibited Con-A-induced TNF-α (A) and IFN-γ (B) production in a dose-dependent manner compared to Con-A-treated cells. The extracellular levels of TNF-α and IFN-γ were measured in culture media using commercial ELISA kits. Results indicated the mean ± SEM of three ndependent experiments. The asterisks show statistically significant values (*p < 0.05, **p < 0.01, and ***p < 0.001).
Effects of osthole on LPS or Con-A-induced TNF-α and IFN-γ mRNA expression
The qPCR was performed to determine whether osthole inhibited the expression of TNF-α and IFN-γ at the transcriptional level. Treatment with LPS increased the expression of TNF-α 7.4-fold compared to the control (Fig. 4A); osthole significantly reduced the expression of LPS-induced TNF-α in a dose-dependent manner. Osthole inhibited LPS-induced TNF-α expression at a dose of 0.1 μM (p < 0.05). Similar inhibitory effects of osthole were observed on Con-A-induced TNF-α expression (Fig. 4B). Treatment with Con-A increased TNF-α mRNA expression 3.6-fold compared to the control; osthole reduced Con-A-induced TNF-α expression in a dose-dependent manner. Osthole inhibited Con-A-induced TNF-α mRNA expression at a dose of 1 μM (p < 0.01). Therefore, osthole inhibited LPS- and Con-A-induced TNF-α mRNA in PBMCs.
Treatment with LPS significantly increased the expression of IFN-γ mRNA by 2.9-fold compared to the control (Fig. 5A). However, treatment with osthole inhibited LPS-induced IFN-γ mRNA expression significantly when compared to the LPS-treated group. Treatment with Con-A increased the expression of IFN-γ mRNA by 24-fold compared to the control (Fig. 5B). However, treatment with osthole diminished Con-A-induced IFN-γ expression in a dose-dependent manner. Osthole inhibited Con-A-induced IFN-γ mRNA expression at a dose of 1 μM (p < 0.05). Osthole only inhibited Con-A-stimulated IFN-γ mRNA expression in PBMCs.
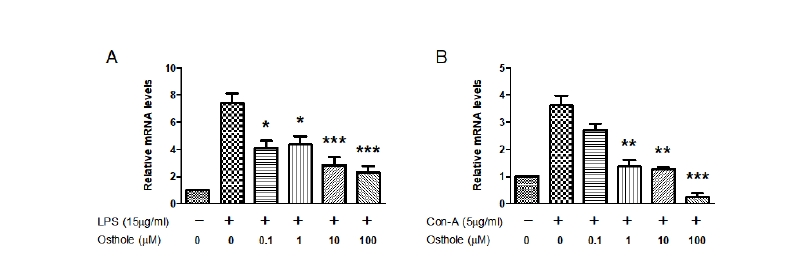
Fig. 4. Effects of osthole on lipopolysaccharide (LPS)- or concanavalin-A (Con-A)-induced tumor necrosis factor-α (TNF-α) gene expression in peripheral blood mononuclear cells (PBMC). Cells were treated with the indicated concentrations of osthole (0, 0.1, 1, 10, and 100 μM) with LPS (15 μg/mL) or Con-A (5 μg/mL) for 6 hours, respectively. Osthole significantly reduced the expression of LPS-induced (A) and Con-A-induced (B) TNF-α in a dose-dependent manner. Results indicated the mean ± SEM of three independent experiments. The asterisks show statistically significant values (*p < 0.05, **p < 0.01, and ***p < 0.001).
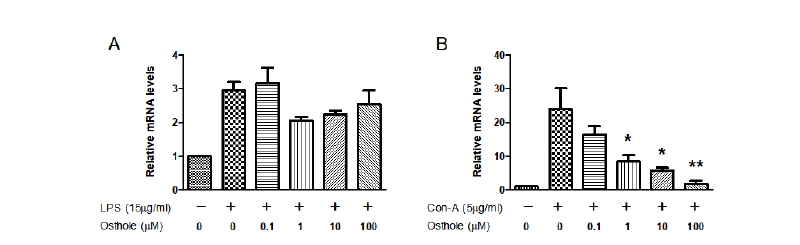
Fig. 5. Effects of osthole on lipopolysaccharide (LPS)- or concanavalin-A (Con-A)-induced interferon-γ (IFN-γ) gene expression in peripheral blood mononuclear cells (PBMC). Cells were treated with the indicated concentrations of osthole (0, 0.1, 1, 10 and 100 μM) with LPS (15 μg/mL) or Con-A (5 μg/mL) for 6 hours, respectively. Treatment with osthole inhibited LPS-induced IFN-γ mRNA expression significantly when compared to the LPS-treated group (A). Treatment with osthole diminished Con-A-induced IFN-γ expression in a dose-dependent manner (B). Results indicated the mean ± SEM of three independent experiments. The asterisks show statistically significant values (*p < 0.05 and **p < 0.01).
Discussion
The present study was performed to assess the pharmacological effects of osthole on the LPS- or Con-A-stimulated production of pro-inflammatory cytokines in bovine PBMCs in vitro. These released endotoxins activate immune cells, endothelial cells, and fibroblasts to stimulate the production and release of various inflammatory mediators including TNF-α and IFN-γ (Abbas and Lichtman, 2012). The activation of cytokines is a key process during the inflammatory reaction (Utsunomiya et al., 1998). These results suggest that osthole is an effective inhibitor of LPS- and Con-A-induced cytokines in PBMCs. Therefore, the inhibitory effects of osthole on the expression of inflammatory mediators comprise one of the mechanisms behind its anti-inflammatory effects, suggesting that osthole might be a potential therapeutic agent for treating inflammatory disease.
Inflammation is a response to injuries related to chemical or microbiological toxins (Guzik et al., 2003). The excess production of pro-inflammatory cytokines is associated with multiple organ dysfunction and mortality (Dinarello, 1996). LPS, the major endotoxin component of Gram-negative bacterial infections, can stimulate systemic inflammatory responses that might lead to shock and multiorgan failure (Bernard et al., 1994). In addition, Con-A treated mice caused T-cell-mediated liver injury. The infiltration of large numbers of inflammatory CD4 + T helper cells into the liver was associated with increased plasma levels of IFN-γ and IL-2, which results in liver damage (Zhang et al., 2010). Therefore, efforts to design treatment strategies have focused on eliminating the potentially detrimental pro-inflammatory mediators.
Osthole, a natural coumarin compound used in traditional Chinese medicines, exerts anti-inflammatory effects, but its effects on cows were unknown. Because TNF-α and IFN-γ are important cytokines in inflammation reactions, we evaluated the effects of osthole on LPS- and Con-A-induced TNF-α and IFN-γ production. Osthole reduced the induced TNF-α and IFN-γ production in a dose-dependent manner in PBMCs. However, inhibitory effects were not seen on LPS-induced IFN-γ production (Fig. 2B). Because LPS did not stimulate the synthesis of IFN-γ, no effects of osthole were observed. Data revealed that the gene and protein expression levels were consistent. A 10-μM dose of osthole significantly protected against inflammation. This suggests that the inhibitory effects of osthole on LPS- or Con-A-induced pro-inflammatory cytokine expression might confer an advantage that protects the host from endotoxic shock.
Many reports have suggested that osthole inhibits inflammatory diseases such as arthritis and hepatitis by modulating the production of inflammatory cytokines, including TNF-α, IFN-γ, and interleukins (Okamoto et al., 2003; Yang et al., 2003; Chiu et al., 2008; You et al., 2009). Both in vitro and in vivo studies have revealed that osthole also exerts anti-inflammatory effects by inhibiting the expression of TNF-α, IL-1β, COX-2, and iNOS (Liao et al., 2010; Li et al., 2012).
This study clearly revealed that osthole inhibited the LPS- or Con-A-stimulated upregulation of pro-inflammatory cytokines in a dose-dependent manner, without causing obvious cytotoxicity. In a bovine model, osthole protected cows from LPS- or Con-A-induced endotoxin shock, possibly by inhibiting the production of pro-inflammatory cytokines, which suggests that osthole may be a potential novel therapeutic agent for the prevention of inflammatory diseases. These data also revealed specific features of the immune responses that increase our knowledge of bovine immunity.
In summary, this study revealed that treating PBMCs with osthole decreased LPS or Con-A-induced pro-inflammatory mediators. Osthole significantly inhibited the mRNA levels secretion of TNF-α and IFN-γ in a dose-dependent manner. Therefore, we conclude that osthole might have the potential to treat patients with, or at risk of, septic shock and other inflammatory diseases. These findings might provide scientific evidence to explain the anti-inflammatory properties of osthole in vitro.
Conclusion
This study revealed that treating PBMCs with osthole decreased LPS or Con-A-induced pro-inflammatory mediators. Osthole significantly inhibited the mRNA levels secretion of TNF-α and IFN-γ in a dose-dependent manner. Therefore, we conclude that osthole might have the potential to treat patients with, or at risk of, septic shock and other inflammatory diseases. These findings might provide scientific evidence to explain the anti-inflammatory properties of osthole in vitro.
Acknwledgements
This work was carried out with the support of "Cooperative Reaserch Program for Agriculture Science & Technology Development (Project title: Genetic variation and gene expression in a parental origin-specific manner for Selection of Hanwoo cows, Project No. PJ01098001)" Rural Development Administration, Republic of Korea and was supported by 2015 PostDoctoral Fellowship Program of National Institute of Animal Science, Rural Development Administration, Republic of Korea.
Authors Information
Seung-Chang Kim, Animal Genomics & Bioinformatics Division, National Institute of Animal Science, Researcher
Seung-Hwan Lee, Division of Animal and Dairy Science, Chungnam National University, Professor
Han-Ha Chai, Animal Genomics & Bioinformatics Division, National Institute of Animal Science, Researcher
Ui-Hyung Kim, Animal Genomics & Bioinformatics Division, National Institute of Animal Science, Researcher
Ki-Yong Chung, Animal Genomics & Bioinformatics Division, National Institute of Animal Science, Researcher
Sun-Sik Jang, Animal Genomics & Bioinformatics Division, National Institute of Animal Science, Researcher
Bong-Hwan Choi, Animal Genomics & Bioinformatics Division, National Institute of Animal Science, Researcher




