Introduction
In the last few decades, effectromics have emerged as a prominent tool for mining effectors from the genomes of pathogens and paving a path for the detection of resistance (R) proteins in plants (Bozkurt et al., 2012). Phytophthora spp. is known for its enormous genome size, where the effectors constitute a major portion of the genome. Effectors are proteins secreted by pathogens to facilitate infection by manipulating host immunity. These effectors facilitate pathogenic invasion in the host and help establish a successful infection. In contrast, effector proteins are recognized by R proteins, which act as the second line of defense, ultimately leading to effector-triggered immunity (ETI) (Tsuda and Katagiri, 2010).
Effectors secreted by Phytophthora spp. can be classified into two main categories, apoplastic and cytoplasmic (Kamoun, 2006), based on their differential localization in the plant cell. Apoplastic effectors are secreted into the plant intercellular space, whereas cytoplasmic effectors are translocated inside plant cells (Kamoun, 2006; Petre and Kamoun, 2014). One important class of cytoplasmic effectors, RXLR effectors, comprises N-terminal regions with a signal peptide followed by a conserved RXLR consensus sequence motif that plays an obscure role in secretion or translocation into the plant cell cytoplasm (Kamoun, 2006; Win et al., 2007; Birch et al., 2008; Arif et al., 2018). The RXLR motif in oomycete effectors is similar to the host translocation signal in malaria parasites, enabling the delivery of effector proteins inside plant cells (Bhattacharjee et al., 2006; Whisson et al., 2007; Dou et al., 2008). RXLR effectors are a subject of effector-assisted breeding research. While RXLR effectors have been mined and reported in large numbers from oomycete pathogen genomes, identifying their biological activities and specific targets inside the cell is still challenging. Among the extensively studied RXLR effectors, P. infestans AVR3a and AVRblb2 are a modular RXLR effector that instigates avirulence activity in potato plants corresponding to the R3a and Rpiblb2 proteins (Armstrong et al., 2005; Oh et al., 2009; Arif et al., 2018). Another added biological role of AVR3a is suppression of cell death induced by Phytophthora infestans (INF1) elicitin (Bos et al., 2006; 2009). Likewise, RXLR effectors produced by Phytophthora sojae and Plasmopara viticola have been reported to suppress Bcl-Associated X (BAX)-triggered programmed cell death (BT-PCD) in Nicotiana benthamiana (Wang et al., 2011). Two oomycete RXLR effectors, PSR1 and PSR2 of P. sojae, were shown to suppress RNA-silencing in plants by inhibiting the biogenesis of small RNAs (Qiao et al., 2013).
In addition to oomycete pathogens, several other plant pathogens, including fungi, bacteria, and nematodes, have also been reported to secrete RXLR-like effectors involved in diverse activities in plant cells. Ustilaginoidea virens effectors have been reported to suppress the Burkholderia glumae-triggered hypersensitive response (HR) in N. benthamiana, and trigger cell death in rice protoplasts (Fang and Tyler, 2016). Effectors reported from Magnaporthe oryzae induced cell death in rice protoplasts (Chen et al., 2013). Several species of nematodes, including Meloidogyne incognita, M. javanica, and Heterodera avenae, have been reported to suppress BAX-triggered PCD (Chen et al., 2013).
Phytophthora capsici is a hemi-biotrophic oomycete plant pathogen with a broad host range, which includes vegetable crops. Globally, P. capsici is known to cause the principal disease of Capsicum annuum and also attacks a broad range of plant species belonging to the Cucurbitaceae, Leguminosae, Solanaceae, and Cruciferae families (Lamour et al., 2012b). In part, this pathogen is difficult to manage due to its production of long-lasting sexual spores, and its tendency to rapidly evolve fungicide resistance (Granke et al., 2012). Over the years, whole-genome sequencing of P. capsici has been performed at various research facilities. Bioinformatics tools have predicted hundreds of effectors in P. capsici genome (Lamour et al., 2012a). These predicted effectors serve as potential targets for identifying related R proteins that mediate effector-triggered breeding.
Moreover, assigning biological roles to the predicted effectors is the focal point of effector-related studies. Here, we characterized the role of two predicted RXLR effectors from P. capsici. Effectors were screened for three different biological activities: (1) suppression of INF1 or BAX-triggered PCD in N. benthamiana, (2) specific induction of HR in three commercial cultivars of C. annum. The PcREK6 and PcREK41 effectors showed HR in chili. PcREK6 fully suppressed PCD triggered by BAX, while PcREK41 partially suppressed PCD triggered by INF1.
Materials and methods
Plant materials and growth conditions
The chili pepper cultivars (Capsicum. annuum cv. Jumbo, Jumping, and Jindaegeon) were provided by the Asia Seed Company (Seoul, Korea). (C. annuum( plants were grown under a 16/8 hr light/dark photoperiod at 25℃. Five-week-old plants were transplanted and grown in individual pots in a growth chamber as described previously (Oh et al., 2010c). Nicotiana benthamiana plants were grown in a plant growth chamber at 25℃. Five-week-old plants were used for agro- infiltration assays.
Cloning of PcREK effector genes
Phytophthora capsici isolate (KACC isolate) was obtained from the Korean Agricultural Culture Collection (KACC). Candidate RXLR effector genes of P. capsici were mined from the database developed by Lamour et al. (2012a). Two non-redundant RXLR effectors were selected, and primer pairs were designed based on the mature region of candidate RXLR effectors (Table 1). RXLR effector genes were amplified from three P. capsici isolates as templates. The PCR products were cloned into the pMD20-T vector (Takara Bio, CA, USA) and sequenced. Sequence similarity searches were performed using standard bioinformatics programs such as BLAST (https://blast.ncbi.nlm.nih.gov/Blast).
Plasmid constructs
RXLR effectors of P. capsici were cloned into a binary potato virus X-based pGR106 vector using ligation-independent cloning (LIC) as described previously (Oh et al., 2010b), and later transformed into E. coli DH5α and Agrobacterium tumefaciens GV3101. The pGR106-dGFP and pGR106-RD2 constructs were used as negative and positive controls, respectively.
Agro-infiltration assays
For HR cell death induction assays, three chili pepper cultivars (C. annuum cv. Jumbo, Jumping, and Jindaegeon) were used. N. benthamiana plants were used for the cell death suppression assay. A. tumefaciens strain GV3101 carrying pGR106-PcREK genes was grown in liquid YEP medium with appropriate antibiotics at 28℃ for 2-days, before centrifugation and resuspension in infiltration medium as previously described (Oh et al., 2010c). We performed a duplicate infiltration assay of the RXLR effector, and observed the cell death response in chili peppers for all effectors. Cell death induction caused by an interaction between the RXLR effector and chili pepper was observed until 10 days after infection (DAI).
For the cell death suppression assays, A. tumefaciens cultures expressing the RXLR effectors or controls were infiltrated into N. benthamiana. One day later, the infiltration sites were challenged with recombinant A. tumefaciens carrying p35S-INF1 and BAX at a final OD600 of 1, as previously described (Bos et al., 2006; 2009). Assays were repeated three times, and infiltrations were made on three leaflets of one plant. The final observations for cell death suppression phenotypes were made at 3 and 4 DAI (Chen et al., 2013).
Reverse transcription polymerase chain reaction (RT-PCR) analysis
RXLR gene expression during infection was determined as previously described (Oh et al., 2010c). cDNA synthesis was performed from 2 μg of total RNA using cDNA synthesis Master Mix (LeGene Biosciences, San Diego, CA, USA) according to the manufacturer’s instructions. The actin gene and P. capsici elongation factor-alpha were used to monitor transcript levels as controls (Oh et al., 2010a). Phenotypic observations were made at 3 - 4 DAI.
Results
Prediction and cloning of RXLR effectors from Phytophthora capsici
We successfully carried out allele mining of RXLR effectors using P. capsici RXLR effector database developed by Lamour et al. (2012a). The pathogenicity of P. capsici isolates towards the chili cultivar C. annuum cv. Chilsungcho (P. capsici-susceptible) was assessed, followed by genomic DNA extraction of P. capsici using the phenol/chloroform method. PCR amplicons of PcREK were cloned into the pGR106 vector, with the backbone of potato virus-X, for high-throughput screening via transient overexpression in plants (Terauchi et al., 2005; Oh et al., 2010c), using the ligation independent cloning (LIC) method. The LIC method allows for rapid cloning, resulting in many PCR amplicons without the use of ligase and restriction enzymes (Oh et al., 2010b).
The identified effectors were named the PcREK (P. capsici RXLR Effectors from Korea isolates) genes. The sequencing results confirmed the effectors with the typical conserved motif RXLR, which is located adjacent to the signal peptide at the N-terminus of the effectors (Table 2). Most of the effectors also had a second conserved motif, namely dEER (Ser/Asp-Glu-Glu-Arg) motif, located downstream of the RXLR motif (Wawra et al., 2012).
Table 2. Amino acid sequences of PcREK6 and PcREK41.

|
|
Underlines indicate the RXLR amino acid sequences. Asterisks are stop codons. PcREK, Phytophthora capsici RXLR effectors from Korea isolate. |
The PcREK effector genes are expressed during infection of chili pepper plants
To validate whether the PcREK effector genes of P. capsici were expressed during infection, RT-PCR was performed using specific primers (Table 1). The expression of 2 PcREK genes for 4 days was examined using chili pepper leaves (cv. Jindaegeon) inoculated with P. capsici isolate (KACC isolate) (Fig. 1). The P. capsici elongation factor 1 alpha (PcEF1a) (Blair et al., 2008), and C. annuum actin (CaActin) gene (Oh et al., 2010a), were used as controls. The constitutively expressed CaActin gene was used to adjust transcription levels. It has been shown that most PcREK effectors are predominantly expressed on days 3 and 4, confirming their expression as effectors of pathogen infection. Two effectors, PcREK6 and PcREK41, were expressed 3 days after inoculation with the pathogen (Fig. 1).
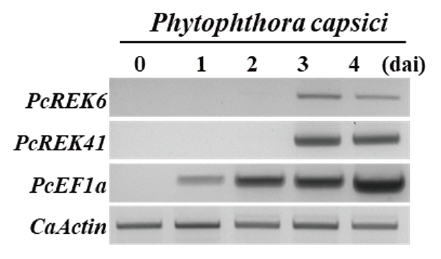
Fig. 1. Reverse transcription polymerase chain reaction (RT-PCR) analysis. RXLR effectors of Phytophthora capsici are expressed during infection of chili pepper. P. capsici KACC isolate was inoculated on chili leaves (Capsicum annuum cv. Jindaegeon). Infected leaves were harvested at 0, 1, 2, 3, 4 days after inoculation (dai) for total RNA extraction. RT-PCR was performed using the P. capsici RXLR effectors from Korea isolate (PcREK) effector specific primers of P. capsici. The constitutive C. annuum Actin (CaActin) and P. capsici elongation factor 1 alpha (PcEF1a) were used as controls.
PcREK genes induce cell death response in chili
Ectopic expression of effector genes in plant cells often leads to macroscopic phenotypes such as cell death, chlorosis, and tissue browning (Kjemtrup et al., 2000; Torto et al., 2003; Haas et al., 2009; Cui et al., 2015; Guo et al., 2020). Agro-infiltration assays were performed to characterize the cell death induction activity of 2 PcREK effectors in chili peppers (Huitema et al., 2004). The results revealed that PcREK6 and PCREK41 genes could trigger cell death response at least in one of the chili pepper cultivars. (Fig. 2C and 2D). We also investigated the frequency of cell death response in chili peppers and rated cell death at the inoculation sites (Fig. 2E). Both the PcREK effectors induced cell death responses in three chili pepper cultivars, but PcREK41 showed the most severe and steady cell death responses. The behavior PcREK41 being consistent with that of the HR phenotypes suggests a possible interaction between the effector and the R protein (resistant protein).
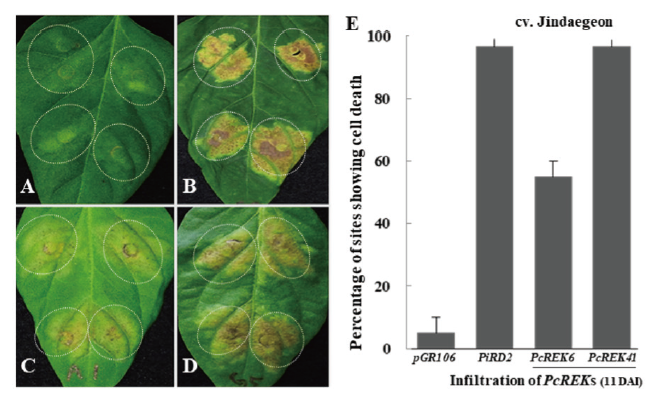
Fig. 2. Cell death screening of RXLR candidate PcREK effectors in chili pepper. Agro-infiltrated effector expressing hypersensitive response (HR) cell death phenotypes in chili pepper cultivar of Jindaegeon. HR phenotypes observed, from mild to strong cell death phenotypes. (A) Negative control; pGR106 vector only. (B) Positive control; PiRD2 an effector from Phytophthora infestans expressing HR symptoms. (C) PcREK6 ; Agro-infiltrated sites showing mild expression of HR. (D) PcREK41 ; Agroinfiltrated sites showing strong expression of HR. (E) HR phenotypic expression measured as percentage cell death at 11 days after infection (DAI). All experiments replicated three times. Error bars indicate the standard deviation. PiRD2, Phytophthora infestansRD2; PcREK, Phytophthora capsici RXLR effectors from Korea isolate.
To confirm the HR cell death response by PcREK41, we conducted agro-infiltration assays comprising of three independent experiments with 20 infiltrations per experiment. Even though Agrobacterium-mediated transient gene expression for chili pepper has relatively low efficiency, infiltration of PcREK41 resulted in more than 90% cell death in the infiltration sites of all chili pepper cultivars at 9 - 11 DAI (Fig. 3A and 3C). The chili pepper plants infiltrated with PcREK41 initiated a cell death response from 3 to 4 DAI. This percentage of cell death response was identical to that of pGR106-PiRD2 effector of P. infestans (Fig. 3A and 3C). These results showed that PcREK41 interacted with the putative R gene of chili pepper to trigger a cell death response.
RT-PCR analysis of PcREK41 confirmed the expression of the relative effector (Fig. 3B). The expression of pGR106-PiRD2 effector genes in P. infestans was examined using chili pepper leaves (Jindaegeon, Jumping, and Jumbo). The constitutively expressed CaActin gene was used to adjust transcription levels. (Fig. 3B). The results showed that expression increased in all three cultivars, and was highest in cv. Jumping and cv. Jindaejeon.
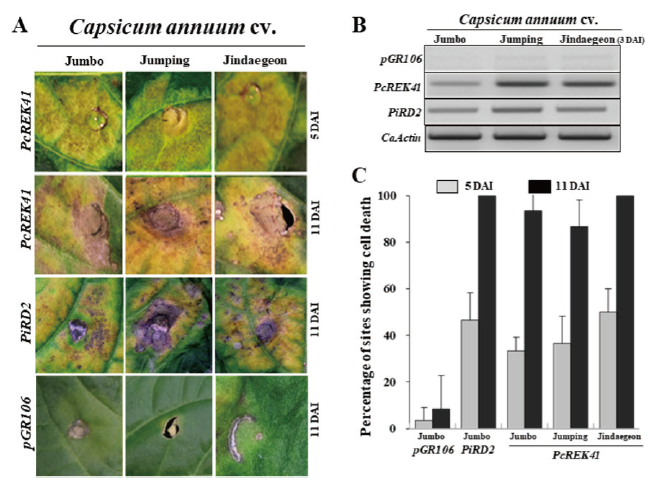
Fig. 3. PcREK41 effectors triggering hypersensitive response (HR) in chili pepper. (A) Shows progression of HR cell death over period of time for PcREK41 in the cultivars of Capsicum annuum cv. Jumbo, Jumping and Jindaegeon. Leaves were photographed at 5 and 11 days after infection (DAI). For control treatments pGR106-vector only and PiRD2 were used. (B) RT-PCR analysis for PcREK41 as confirmation for the expression of relative effectors. Capsicum annuum Actin (CaActin) was used as internal control for the equal quantification of RNA among the samples. (C). HR phenotypic expression measured as percentage cell death over span of 0 to 11 DAI. The percentage cell death gradually elevated, maximized on 11 DAI. All experiments replicated three times at least. Error bars indicate the standard deviation. PiRD2, Phytophthora infestans RD2; PcREK, Phytophthora capsici RXLR effectors from Korea isolate.
Effectors suppress cell death in N. benthamiana
To characterize the cell death suppression assays of the two PcREK effectors in chili peppers, BAX-induced Agro-infiltration assays were performed. BAX, a cell death-promoting member of the Bcl-2 family of proteins, triggers cell death when expressed in plants from a tobacco mosaic virus vector. The cell death-promoting function of BAX in plants correlated with the accumulation of the defense-related protein PR-1, suggesting that BAX activates an endogenous cell death pathway in plants. (Lacomme and Santa, 1999). Our results showed that PcREK6 could completely suppress BAX-induced cell death when co-infiltrated with N. benthamiana (Fig. 4), while PcREK41 partially suppressed BAX-induced cell death, as indicated by weaker necrosis. These results showed that PcREK6 suppressed BAX-triggered cell death response (Fig. 4).
To better understand the suppression of cell death by the two PcREK effectors, we performed INF1-induced Agro-infiltration assays. In the case of INF1 induced cell death suppression assay, the PcRDK6 effector was unable to suppress necrosis triggered by the expression of INF1. However, PcREK41 could partially suppress the INF1 triggered cell death (Fig. 5). These results suggest that the PcREK41 effector may play a role in suppressing host immunity through alternate pathways (other than INF1 related PTI pathways), as indicated by the weaker necrosis resulting in co-infiltration of effectors with INF1 (Fig. 5).
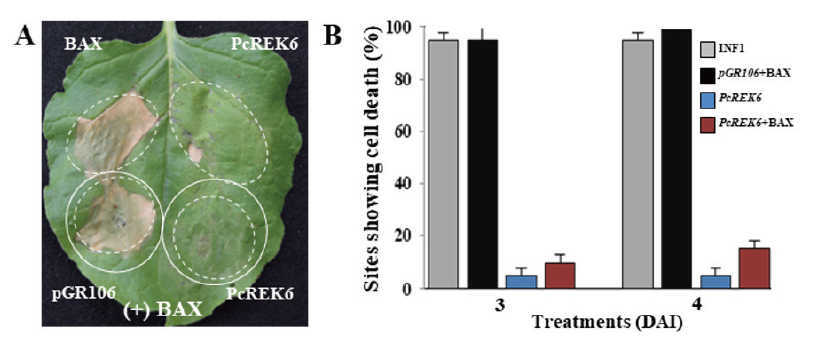
Fig. 4. PcREK6 suppresses BAX induced cell death in N. benthamiana. (A) Symptoms of infiltration sites co-expressing PcREK6 or pGR106 (vector only) with BAX (+BAX). Where PcREK6 fully suppresses the BAX induced cell death. (B) Percentages of infiltration sites showing BAX induced cell death upon co-expression of PcREK6 with BAX at 3 and 4 days after infection (DAI). Error bars indicate the standard deviation. BAX, Bcl-Associated X; PcREK, Phytophthora capsici RXLR effectors from Korea isolate; INF1, Phytophthora infestans elicitin.
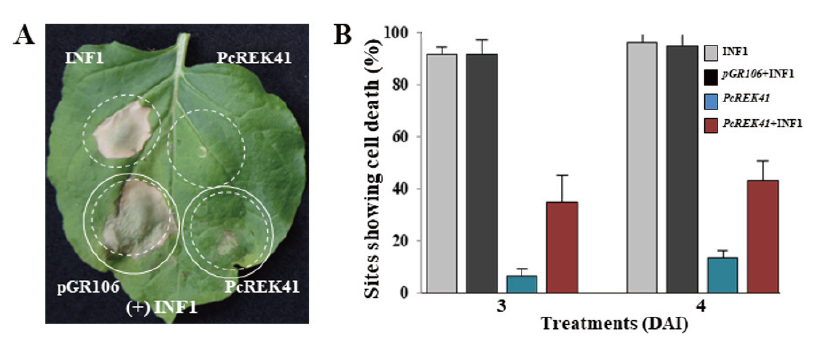
Fig. 5. PcREK41 suppresses INF1 induced cell death in N. benthamiana. (A) Symptoms of infiltration sites co-expressing PcREK41 or pGR106 (vector only) with INF1 (+INF1). Where PcREK41 partially suppresses the INF1 induced cell death. (B) Percentages of infiltration sites showing INF1 induced cell death upon co-expression of PcREK41 with INF1 at 3 and 4 days after infection (DAI). Error bars indicate the standard deviation. INF1, Phytophthora infestans elicitin; PcREK, Phytophthora capsici RXLR effectors from Korea isolate.
Discussion
The filamentous oomycete Phytophthora capsici is a highly destructive vegetable pathogen that causes severe economic losses worldwide. Phytophthora spp., including P. capsici, encode hundreds of RXLR effectors in their genomes (Whisson et al., 2007; Win et al., 2007; Birch et al., 2008). These encoded candidate proteins manipulate host cells to establish a successful infection. In contrast, the RXLR effectors can also trigger ETI. Thus, it is crucial to understand the molecular mechanisms of P. capsici effectors that play an important role in infection. In this study, two candidate RXLR effectors were predicted from the P. capsici genome for their distinct biological activities in a host cell. First, we predicted two candidate RXLR effectors using the bioinformatics database developed by Lamour et al. (2012a), from the genome sequence of P. capsici (Lamour et al., 2012a). We obtained two primer pairs from the P. capsici isolate, and generated a library of RXLR effector clones. Two candidate effectors were found to be secreted during infection, as confirmed by RT-PCR, suggesting the efficacy of cDNA even in the absence of a whole-genome sequence (Torto et al., 2003; Tian et al., 2004; Liu et al., 2005).
RXLR effectors are known for their avirulence (Avr) activity against the resistant host with associated R genes, and it is one of the most important biological activities attributed to RXLR effectors. To validate the interaction between the RXLR effectors of P. capsici and chili pepper, we inoculated two PcREK effectors into three chili pepper cultivars using potato virus X (PVX)-Agro infiltration methods. Our results revealed that the two effectors were able to induce cell death in at least one of the chili cultivars. However, it cannot be confirmed that HR induction was solely based on an interaction between the RXLR effector of P. capsici and the putative R gene of chili pepper, and not some other underlying biological phenomenon. Avr effectors from bacteria, extracellular fungi, and oomycetes have been well known for many years (Espinosa and Alfano, 2004). Avr genes were initially isolated from oomycete pathogens: Avrlb-1 from Phytophthora sojae, Avr3a from P. infestans, and ATR13 and ATRINaWsB from Hyaloperonospora arabiaopsiáis, the downy mildew of Arabidopsis, and are known to alter host immunity, resulting in tissue necrosis, browning, and chlorosis (Allen et al., 2004; Armstrong et al., 2005; Rehmany et al., 2005). The induction of HR elicited by the two PcREK effectors demonstrates the effector activity of this protein, further indicating its potential to be recognized by plant R genes.
Most of the Avr and other RXLR proteins identified so far show no homology to any proteins of known biological activity, except for some serine proteases. Hence, the data regarding their roles in infection is currently insuffecient. However, one known activity, namely the suppression of PTI, has emerged as an important activity of the effectors (Block et al., 2008; Hogenhout et al., 2009). The well-known RXLR effector P. infestans Avr3a prevents cell death induced by the INF1 elicitor (Bos et al., 2006). Our results also indicated that two PcREK effectors could partially suppress cell death induced by INF1 and BAX in N. benthamiana. While we performed experiments in the non-host N. benthamiana, previous studies have revealed that these effectors retain their ability to suppress or induce cell death in both non-host and host plants. For instance, Avh172 and Avh6 of P. sojae have been shown to suppress ETI in non-host N. benthamiana as well as in host soybean (Wang et al., 2011). In our assay, PcREK6 fully suppressed BAX triggered cell death (Fig. 4), whereas PcREK41 partially suppressed it. In contrast, PcREK41 partially suppressed INF1 triggered cell death (Fig. 5). In brief, PcREK41 showed consistently positive results in all screening assays, including in HR and cell death suppression assays. Further work on PcREK6 and PcREK41 genes is needed to analyze the mode of action and the correlation between the virulence function and the interaction of the two effectors with as yet unknown proteins in planta.
Conclusion
In this study, we successfully cloned two RXLR genes into PVX-based pGR106 vector, using ligation-independent cloning. The screening assay revealed that PcREK6 and PCREK41 genes were involved in HR cell death phenotypes. PcREK6 and PcREK41 genes encode the N-terminal conserved RXLR-DEER motif and signal peptide sequences. Cell death suppression assays in N. benthamiana revealed that PcREK6 fully suppressed PCD triggered by BAX, while PcREK41 partially suppressed PCD triggered by INF1.
Acknowledgments
We thank to the Asia seed Company (Seoul, Korea) for providing the chili pepper cultivars (Jumbo, Jumping, and Jindaegeon). We also thank Korean Agricultural Culture Collection for providing the Phytophthora. capsici isolate. This work was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through Crop Viruses and Pests Response Industry Technology Development Program, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) (Project No. 120086052SB010).





